Cross-project comparisons for disease markers¶
GMrepo currently supports two types cross-project comparisons for disease markers, see details below
Cross-project and phenotype comparison for a maker taxon¶
This purposes of this comparison are to show if a specific taxon:
- is unique to a disease, or shared by multiple diseases,
- has the same trend across diseases (e.g., always enriched in disease- or health- associated samples), or different trends in different diseases,
- has the same trend in multiple project of the same disease, or conflicting trends in different projects of the same diseases.
Here we use two examples to explain what the above purposes mean.
1. Fusobacterium nucleatum ¶
F. nucleatum is a known marker for colorectal cancer (CRC). In GMrepo, we show that it is also a marker for multiple diseases, including:
- Crohn Disease,
- COVID-19,
- Cardiovascular Diseases,
- Inflammatory Bowel Diseases, and
- Liver Cirrhosis

In addition, it shows consistent trends as a disease-enriched marker:
- not only across all above diseases,
- but also across multiple projects of the same diseases, including
- Crohn Diseases, and
- Colorectal Neoplasms (CRC).
2. Prevotella copri ¶
P. copri is also a marker species for multiple diseases:

However, it shows inconsistent trends not only in different diseases (e.g., enriched in Adenoma but depleted in Crohn Diseases), but also in different projects in the same disease (e.g., in Autism Spectrum Disorder and Colorectal Neoplasms).
Marker taxa of a disease across multiple projects¶
In GMrepo , a disease could be covered by multiple datasets/projects. To facilitate cross-project comparisons of the identified markers within each project, a dedicated page for each phenotype pair (e.g., health versus liver cirrhosis, or adenoma versus colorectal cancer) is available to systematically show the consistent and non-consistent disease-associated microbial markers across datasets.
Users can check the Phenotype comparisons page for all available phenotype pairs.
Below we will use two example to show consistent and inconsistent disease markers across projects.
1. Colorectal Newplasms (CRC) ¶
In addition to details of the related projects and a data table of identified marker taxon for CRC (as compared with healthy controls), a tile view is used to visualize all markers stratified by projects:
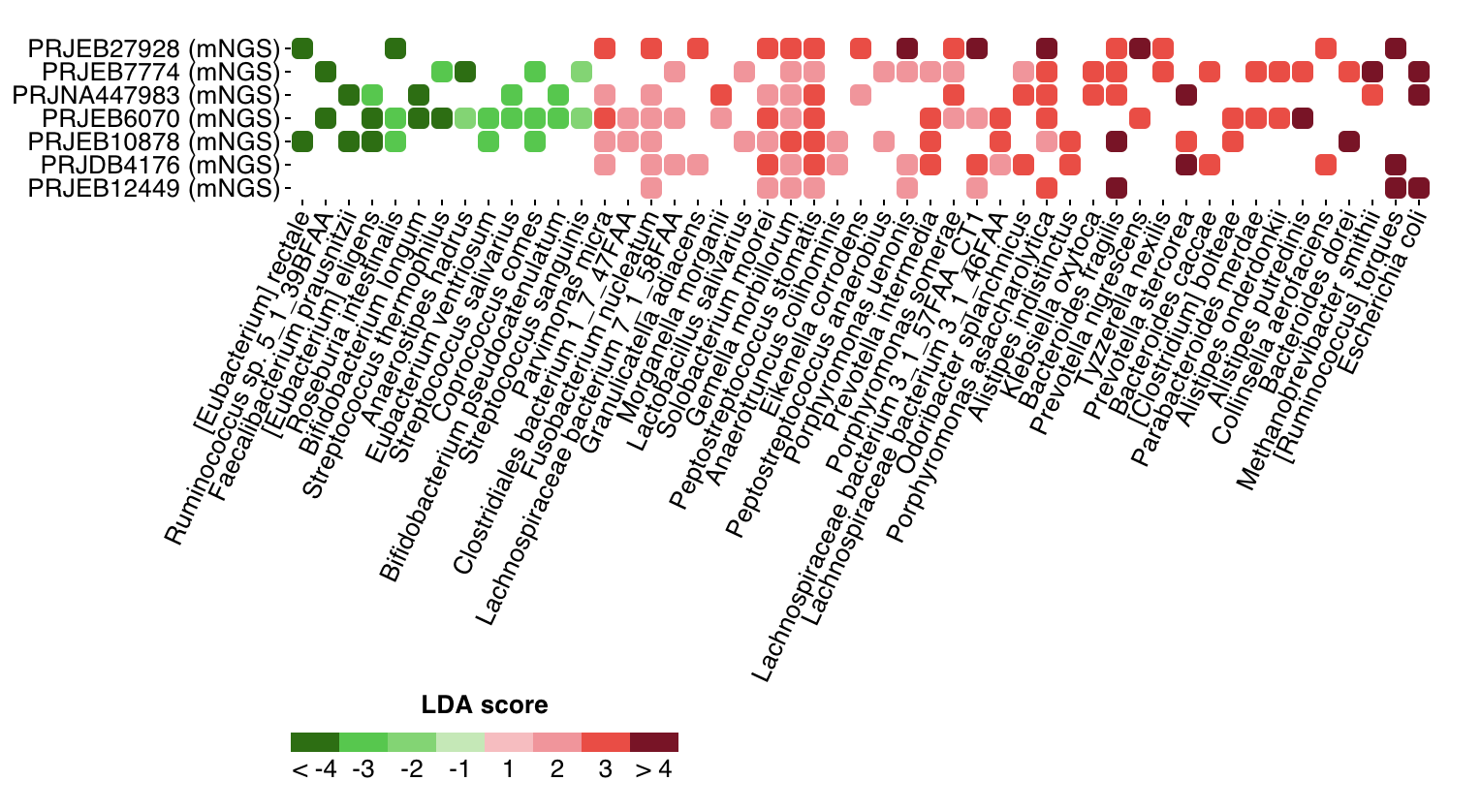
Shown here are microbial markers across the seven projects; it is evident that disease-enriched markers are consistent across projects, including known CRC marker species Fusobacterium nucleatum, Parvimonas micra and Gemella morbillorum.
2. Autism Spectrum Disorder (ASD) ¶
Shown below are markers of ASD (as compared with healthy controls) in 9 projects:
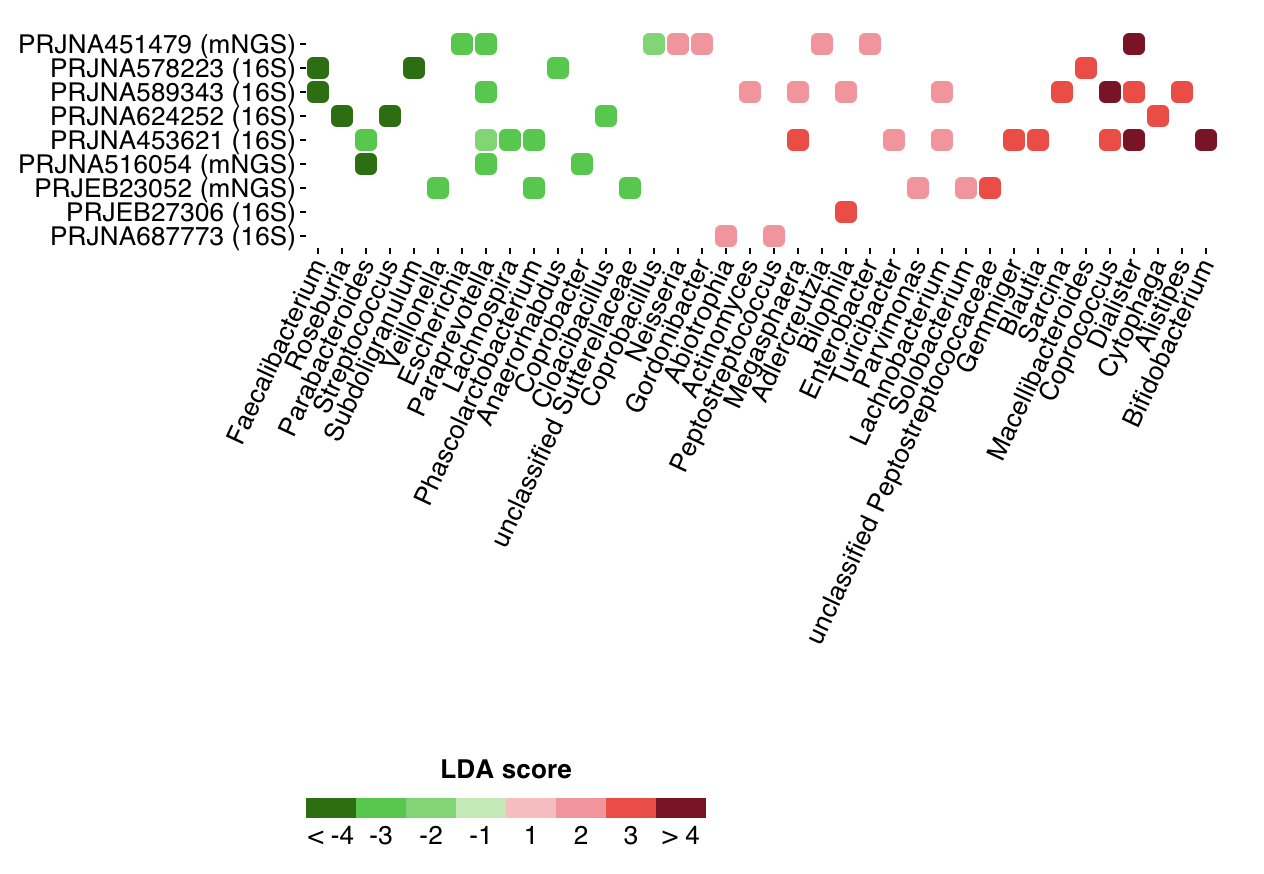
only very few markers can be found in mulple projects.